Expression of Phytase gene in transgenic maize with seed-specific promoter 27-kDa γ Zein and constitutive promoter CaMV 35S
Ririn Septina Arthasari(1), Rani Agustina Wulandari(2*), Panjisakti Basunanda(3)
(1) Department of Agronomy, Faculty, of Agriculture Universitas Gadjah Mada Jln. Flora no. 1, Bulaksumur, Sleman, Yogyakarta 55281, Indonesia
(2) Department of Agronomy, Faculty, of Agriculture Universitas Gadjah Mada Jln. Flora no. 1, Bulaksumur, Sleman, Yogyakarta 55281, Indonesia
(3) Department of Agronomy, Faculty, of Agriculture Universitas Gadjah Mada Jln. Flora no. 1, Bulaksumur, Sleman, Yogyakarta 55281, Indonesia
(*) Corresponding Author
Abstract
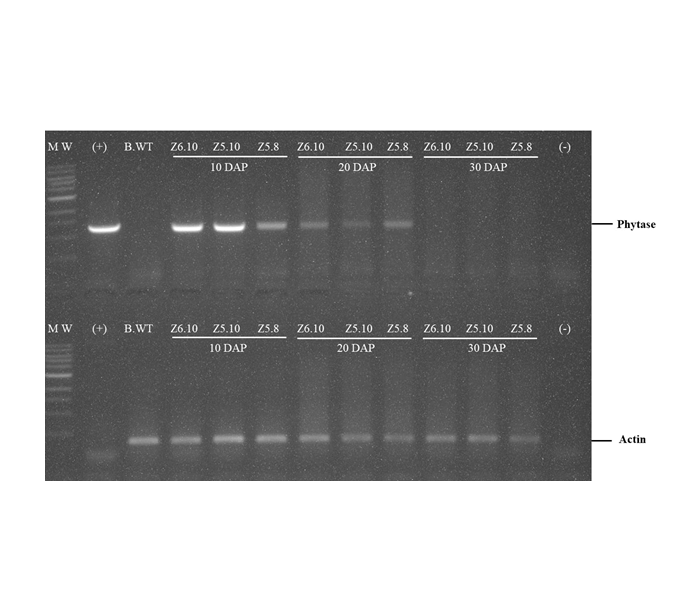
Phytase enzymes are applied to animal feed to help animals absorb more nutrients. The use of feed raw materials containing phytase enzymes is expected to reduce the cost of animal feed production. Efforts to increase the phytase content in maize were carried out by improving genetics, in the way of assembling transgenic plants containing high phytase content. The 27-kDa γ Zein promoter is a specific promoter that expresses genes in caryopsis, and promoter CaMV 35S is a constitutive promoter that controls gene expression in all tissues and generally does not depend on the growth phase. Transgenic maize was transformed using Agrobacterium tumefacien infection method on maize B104. The reverse transcriptase polymerase chain reaction (RT-PCR) approach was used to examine the expression of phytase genes in leaves, roots, and caryopsis was done 10, 20, and 30 days after pollination (DAP). The phytase enzyme activity test was also carried out by using the colorimetric phosphomolybdate analysis method to see the phytase enzyme activity in unit µg-1. The results showed that the phytase gene in transgenic plants with the 27-kDa γ Zein promoter was highly expressed in maize caryopsis, but in line Z6.10 was also expressed in leaves, while in the CaMV 35S promoter the phytase gene was only expressed on the leaves. Phytase enzyme activity showed that transgenic maize was higher than non-transgenic maize.
Keywords
Full Text:
PDFReferences
Abid, N., Khatoon, A., Maqbool, A., Irfan, M., Bashir, A., Asif, I., Shahid, M., Saeed, A., Brinch-Pedersen, H., and Malik, K. A. (2017). Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transgenic Research, 26(1), pp. 109–122.
Bae, H.D., Yanke, L.J., Cheng, K.J., and Selinger, L.B. (1999). A novel staining method for detecting phytase activity. Journal of Microbiological Methods, 39(1), pp 17–22.
Bak, A. and Emerson, J.B. (2020). Cauliflower mosaic virus (CaMV) biology, management, and relevance to GM plant detection for sustainable organic agriculture. Frontiers in Sustainable Food Systems, 4(21), pp. 1–8.
Belgaroui, N., Zaidi, I., Farhat, A., Chouayekh, H., Bouain, N., Chay,S., Curie, C., Mari, S., Masmoudi, K., Davidian, J.C., Berthomieu, P., Rouached, H., and Hanin, M. (2014). Over-expression of the bacterial phytase US417 in Arabidopsis reduces the concentration of phytic acid and reveals its involvement in the regulation of sulfate and phosphate homeostasis and signaling. Plant and Cell Physiology, 55(11), pp. 1912–1924.
Belgaroui, N., Lacombe, B., Rouached, H., and Hanin, M. (2018). Phytase overexpression in Arabidopsis improves plant growth under osmotic stress and in combination with phosphate deficiency article. Scientific Reports, 8(1), pp. 1–12
Borlini, G., Rovera, C., Landoni, M., Cassani, E., and Pilu, R. (2019). lpa1-5525: A New lpa1 mutant isolated in a mutagenized population by a novel non-disrupting screening method. Plants (Basel), 8(7), pp. 209
Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, pp. 248–254.
Chen, R., Guangxing, X., Ping, C., Bin,Y., Wenzhu, Y., Qianli, M., Yunliu, F., Zuoyu, Z., Mitchell, C. T., and Jinrui, S.(2008). Transgenic maize plants expressing a fungal phytase gene. Transgenic Research, 17(4), pp. 633-643
Chiera, J.M., Finer, J.J., and Grabau, E.A. (2004). Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Molecular Biology, 56(6), pp. 895–904.
Corrêa, T.L.R., and de Araújo, E.F. (2020). Fungal phytases: from genes to applications. Brazilian Journal of Microbiology, 51, pp. 1009-1020
Coulibaly, A., Brou, K., and Jie, C. (2011). Phytic acid in cereal grains: structure, healthy or harmful ways to reduce phytic acid in cereal grains and their effects on nutritional quality. American Journal of Plant Nutrition and Fertilization Technology, 1, pp. 1-22.
Dersjant, L.Y., Awati, A., Schulze, H., and Partridge, G. (2015). Phytase in non-ruminant animal nutrition: a critical review on phytase activities in the gastrointestinal tract and influencing factors. J. Sci. Food Agric., 95(5), pp. 878–896
Erpel, F., Restovic, F., and Arce-Johnson, P. (2016). Development of phytase expressing Chlamydomonas reinhardtii for monogastric animal nutrition. BMC Biotechnol., 16(29), pp. 1-7
Frame, B.R., Marcy, M., Rosemarie, S., and Wang, K. (2011). Genetic transformation using maize immature zygotic embryos. Plant embryo culture: methods and protocols. Methods Molecular Biology, 710, pp. 327-341
Geetha, S., Beslin, J., Joshi,K., Kumar, K., Arul, L., Kokiladevi, E., Balasubramanian P., and Sudhakar D. (2019). Genetic transformation of tropical maize (Zea mays L.) inbred line with a phytase gene from Aspergillus niger. 3 Biotech, 9(6), pp. 1–10.
Gontia, I., Tantwai, K., Rajput, L. P. S., and Tiwari, S. (2012). Transgenic plants expressing phytase gene of microbial origin and their prospective applications as feed. Food Technol., Biotechnol., 50(1), pp. 3–10
Guo, X., Yuan, L., Chen, H., Sato, S.J., Clemente, T.E., and Holding, D.R. (2013). Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm. Plant Physiol., 162(3), pp. 1359–1369.
Joshi, J.B., Geetha S., Singh, B., Kumar, K.K., Kokiladevi, E., Arul, L., Balasubramanian, P., and Sudhakar, D. (2015). A maize α-zein promoter drives an endosperm-specifc expression of transgene in rice. Physiol. Mol. Biol. Plants, 21(1), pp. 35–42
Koramutla, M.K., Bhatt, D., Negi, M., Venkatachalam, P., Jain, P.K., and Bhattacharya, R. (2016). Strength, stability, and cis-motifs of in silico identified phloem-specific promoters in Brassica juncea (L.). Frontiers in Plant Science, 7, pp. 1–12.
Lei, X.G., Weaver, J.D., Mullaney, E.U.A., and Azain, M.J. (2013). Phytase a new life for an old enzyme. Annual Rev. Anim. Biosci., 1, pp. 283–309
Moeller, L., Gan, Q., and Wang, K. (2009). A bacterial signal peptide is functional in plants and directs proteins to the secretory pathway. J. Exp. Bot., 60(12), pp. 3337–3352
Nahampun, H.N., Lee, C.J., Jane, J.L., and Wang, K. (2013). Ectopic expression of bacterial amylopullulanase enhances bioethanol production from maize grain. Plant Cell Rep., 32(9), pp. 1393-405
Pawlowski, P. and Somers, A. (1996). Transgene Inheritance in plant genetically engineered by microprojectile bombardment. Mol. Biotechnol., 6(1), pp. 17-30
Reddy, C.S., Vani, K., Pandey, S., Vijaylakshmi, M., Reddy, P.C.O., and Kaul, T. (2013). Manipulating microbial phytases for heterologous expression in crops for sustainable nutrition. Ann. Plant Sci., 2, pp. 436–454
Richardson, A.E., Hadobas, P.A., and Hayes, J. (2001). Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J., 25(6), pp. 641–649
Selle, P.H., and Ravindran. (2007). Phytate-degrading enzymes in pig nutrition. Livestock Science, 113(2-3), pp. 99–122
Singh, B. and Satyanarayana, T. (2011). Microbial phytases in phosphorous acquisition and plant growth promotion. Physiol. Mol. Biol. Plants, 17(2), pp. 93–103
Singh, N., Sonia, K., Kanu, P., Rajneesh, J., and Rakesh Y. (2018). Phytase: The feed enzyme, an overview. In: Gahlawat, S., Duhan, J., Salar, R., Siwach, P., Kumar, S., Kaur, P. (eds)., Advances in Animal Biotechnology and its Applications, 1st ed. Singapore: Springer, pp. 269-327
Sunilkumar, G., LeAnne, M., Emily, L.F., Chandrakanth, E., and Keerti, S. R. (2002). Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Molecular Biology, 50(3), pp. 463–479.
Terada, R. and Shimamoto, K. (1990). Expression of CaMV 35S-GUS gene in transgenic rice plants. MGG Molecular & General Genetics, 220(3), pp. 389–392.
Wang, Y., Ye, X., Ding. G., and Xu, F. (2013). Over expression of phyA and appA genes improves soil organic phosphorus utilization and seed phytase activity in Brassica napus. PLoS One, 8(4), e60801
Woo, Y.M., Hu, D.W., Larkins, B.A. and Jung. R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell, 13(10), pp. 2297–2317.
Woyengo, T.A., and Nyachoti, C.M. (2013). Review: Anti-nutritional effects of phytic acid in diets for pigs and poultry current knowledge and directions for future research. Can. J. Anim. Sci., 93, pp. 9-21.
Yu, S., Cowieson, A., Gilbert, C., Plumstead, P., and Dalsgaard, S. (2012). Interactions of phytate andmyoinositol phosphate esters (IP1-5) including IP5 isomers with dietary protein and iron and inhibition of pepsin. J. Anim. Sci., 90(6), pp. 1824–1832.
Article Metrics
Refbacks
- There are currently no refbacks.
Ilmu Pertanian (Agricultural Science) ISSN 0126-4214 (print), ISSN 2527-7162 (online) is published by Faculty of Agriculture Universitas Gadjah Mada collaboration with Perhimpunan Sarjana Pertanian Indonesia (PISPI) and licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.












_2025_-_kecil_.png)
_2024_kecil_2.png)
